Archaea
The Archaea (or Archea) are a group of single-celled organisms. The name comes from Greek αρχαία, "old ones". They are a major division of living organisms.
| Archaea | |
|---|---|
| |
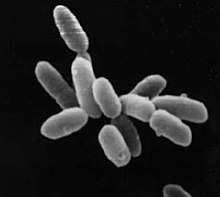
| Halobacterium sp. strain NRC-1, each cell about 5 μm long | |
| Scientific classification | |
| Domain: | Archaea Woese, Kandler & Wheelis, 1990 |
| Kingdoms[3] and phyla[4] | |
| |
| Synonyms | |
| |

Archaea are tiny, simple organisms. They were originally discovered in extreme environments (extremophiles), but are now thought to be common to more average conditions. Many can survive at very high (over 80 °f) or very low temperatures, or highly salty, acidic or alkaline water. Some have been found in geysers, black smokers, oil wells, and hot vents in the deep ocean. Recent research has found ammonia-eating archaea in soil and seawater.
In the past they had been classed with bacteria as prokaryotes (or Kingdom Monera) and named archaebacteria, but this is a mistake.[5] The Archaea have an independent evolutionary history and show many differences in their biochemistry from other forms of life. They are now classified as a separate domain in the three-domain system. In this system, the three distinct branches of evolutionary descent are the Archaea, Bacteria and Eukaryota.
Archaea are, like bacteria, prokaryotes: single-celled organisms that do not have nuclei and cell organelles of the eukaryote type.
Comparison to other domains
changeThe following table compares some major characteristics of the three domains, to illustrate their similarities and differences.[6] Many of these characteristics are also discussed below.
| Property | Archaea | Bacteria | Eukarya |
|---|---|---|---|
| Cell membrane | Ether-linked lipids, pseudopeptidoglycan | Ester-linked lipids, peptidoglycan | Ester-linked lipids, various structures |
| Gene structure | Circular chromosomes, similar translation and transcription to Eukarya | Circular chromosomes, unique translation and transcription | Multiple, linear chromosomes, similar translation and transcription to Archaea |
| Internal cell structure | No membrane-bound organelles (but questioned:[7]) or nucleus | No membrane-bound organelles or nucleus | Membrane-bound organelles and nucleus |
| Metabolism[8] | Various, with methanogenesis unique to Archaea | Various, including photosynthesis, aerobic and anaerobic respiration, fermentation, and autotrophy | Photosynthesis, cellular respiration and fermentation |
| Reproduction | Asexual reproduction, horizontal gene transfer | Asexual reproduction, horizontal gene transfer | Sexual and asexual reproduction |
= Interesting facts about archaea:[9][10][11]
- No archaean species can do photosynthesis.[12]
- Archaea only reproduce asexually.[13]
- Archaea show high levels of horizontal gene transfer between lineages.[14][15]
- Many archaea live in extreme environments.
- Unlike bacteria, no archaea produce spores.
- Archaea are common in the ocean, and especially in the plankton. They make up to 20% of all microbial cells in the ocean.[16]p475
- Carl Woese discovered the Archaea in 1978.
Related pages
changeFurther reading
change- Barry E.R. & Bell S.D. 2006. DNA replication in the Archaea. Microbiology and molecular biology reviews (MMBR) '70, 876-887.
- Kelman L.M. & Kelman Z. 2003. Archaea: An archetype for replication initiation studies? Molecular microbiology, 48, 605-615.
Other websites
change- Archaea -Citizendium
References
change- ↑ 1.0 1.1 "Taxa above the rank of class". List of Prokaryotic names with Standing in Nomenclature. Retrieved 8 August 2017.
- ↑ Cavalier-Smith, T. (2014). "The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life". Cold Spring Harb. Perspect. Biol. 6 (9): a016006. doi:10.1101/cshperspect.a016006. PMC 4142966. PMID 25183828.
- ↑ Petitjean C, Deschamps P, López-García P, Moreira D (December 2014). "Rooting the domain archaea by phylogenomic analysis supports the foundation of the new kingdom Proteoarchaeota". Genome Biology and Evolution. 7 (1): 191–204. doi:10.1093/gbe/evu274. PMC 4316627. PMID 25527841.
- ↑ "NCBI taxonomy page on Archaea".
{{cite journal}}: Cite journal requires|journal=(help) - ↑ Pace NR (May 2006). "Time for a change". Nature. 441 (7091): 289. Bibcode:2006Natur.441..289P. doi:10.1038/441289a. PMID 16710401. S2CID 4431143.
- ↑ Information is from Willey JM, Sherwood LM, Woolverton CJ. Microbiology 7th ed. (2008), Ch. 19 pp. 474–475, except where noted.
- ↑ Thomas Heimerl; et al. (13 June 2017). "A Complex endomembrane system in the Archaeon Ignicoccus hospitalis tapped by Nanoarchaeum equitans". Frontiers in Microbiology. 8: 1072. doi:10.3389/fmicb.2017.01072. PMC 5468417. PMID 28659892.
- ↑ Jurtshuk, Peter (1996). "Bacterial Metabolism". Medical Microbiology (4th ed.). Galveston (TX): University of Texas Medical Branch at Galveston. ISBN 9780963117212. Retrieved 5 November 2014.
- ↑ Howland, John L. (2000). The surprising Archaea: discovering another domain of life. Oxford: Oxford University Press. ISBN 0-19-511183-4.
- ↑ Garrett RA, Klenk H (2005). Archaea: evolution, physiology and molecular biology. WileyBlackwell. ISBN 1-4051-4404-1.
- ↑ Schaechter, M (2009). Archaea (overview) in The desk encyclopedia of microbiology, 2nd edition. San Diego and London: Elsevier Academic Press. ISBN 978-0-12-374980-2.
- ↑ Schäfer G. et al 1999. Bioenergetics of the Archaea. Microbiol. Mol. Biol. Rev. 63 (3): 570–620. PMID 10477309.
- ↑ de Queiroz K (2005). "Ernst Mayr and the modern concept of species". Proc. Natl. Acad. Sci. U.S.A. 102 (Suppl 1): 6600–7. Bibcode:2005PNAS..102.6600D. doi:10.1073/pnas.0502030102. PMC 1131873. PMID 15851674.
- ↑ Eppley JM, Tyson GW, Getz WM, Banfield JF (2007). "Genetic exchange across a species boundary in the archaeal genus ferroplasma". Genetics. 177 (1): 407–16. doi:10.1534/genetics.107.072892. PMC 2013692. PMID 17603112.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Papke RT, Zhaxybayeva O, Feil EJ, Sommerfeld K, Muise D, Doolittle WF (2007). "Searching for species in haloarchaea". Proc. Natl. Acad. Sci. U.S.A. 104 (35): 14092–7. Bibcode:2007PNAS..10414092P. doi:10.1073/pnas.0706358104. PMC 1955782. PMID 17715057.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ DeLong EF, Pace NR (2001). "Environmental diversity of bacteria and archaea". Syst. Biol. 50 (4): 470–8. doi:10.1080/106351501750435040. PMID 12116647. Full text: [1]