Antibody
Antibodies (also called immunoglobulins) are large Y-shaped proteins that can stick to the surface of bacteria and viruses. They are found in the blood or other body fluids of vertebrates. Antibodies are the key element in the adaptive immune system.

2. Fragment crystallizable region
3. Heavy chain (blue) with one variable (VH) domain followed by a constant domain (CH1), a hinge region, and two more constant (CH2 and CH3) domains.
4. Light chain (green) with one variable (VL) and one constant (CL) domain
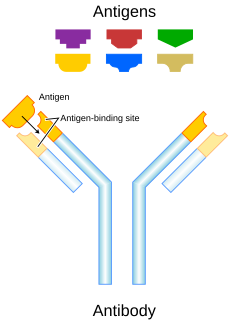
5. Antigen binding site (paratope)
6. Hinge regions
The antibody recognizes a unique part of the foreign target called an antigen.[1][2] Each tip of the "Y" of an antibody contains a structure (like a lock) that fits one particular key-like structure on an antigen. This binds the two structures together.
Using this binding mechanism, an antibody can tag a microbe or an infected cell for attack by other parts of the immune system, or can neutralize its target directly.[3] The production of antibodies is the main function of humoral immunity.[4][5]
Each antibody is different. They are all designed to attack only one kind of antigen (in practice, this means virus or bacteria). For instance, an antibody designed to destroy smallpox is unable to hit the bubonic plague or the common cold.
Though the general structure of all antibodies is very similar, that small region at the tip of the protein is extremely variable. This allows millions of antibodies with different tip structures to exist. Each of these variants can bind to a different antigen.[1] This enormous diversity of antibodies allows the immune system to recognize an equally wide variety of antigens.[6]
Relationship Between White Blood Cells and Antibodies
changeThere are two types of white blood cells: Phagocytes and Lymphocytes.
Phagocytes are a type of white blood cell. Once the antibodies attach to the pathogen the phagocytes bind to and engulf the pathogens. Once the pathogen is surrounded, the phagocyte then breaks down the cell using enzymes in its body. Phagocytes rely on antibodies to flag down pathogens in the blood system.
The other type of white blood cell is called a Lymphocyte. These white blood cells are the ones that are in charge of producing antibodies. The Lymphocytes can detect what pathogen is in your body at that given moment and the Lymphocyte will produce a specific type of antibody to specifically attack that pathogen. These antibodies help group pathogens together into one mass so that the phagocytes can just engulf them whole, rather than going around and individually finding pathogens. They produce antibodies that are specific to the pathogen. It takes a while for lymphocytes to produce these specific antibodies. In addition, some lymphocytes produce toxins that counteract the effect of pathogenic exotoxins in the body.[7]
Immunoglobulin diversity
changeBasic issue
changeAlthough a huge variety of different antibodies is made in a single individual, the number of genes available to make these proteins is limited by the size of the genome.
There are a vast number of microbe strains, and so vertebrates need millions of different antibodies.[8] Actually, humans generate about 10 billion different antibodies, each capable of binding a distinct antigen site.[9] This must be done with a very much smaller number of genes: the total human genome has only about 20,000 genes.
Several complex genetic mechanisms have evolved. These allow vertebrate B cells to generate a huge pool of antibodies from a relatively small number of antibody genes.[10] The full details are not presented here, just a summary.
The variety of antibodies is got by combining segments from a pool of genes in many different ways. Then, hyper-mutations occur in the binding site area of the antibody gene. This creates further diversity.[11][12]
Heavy chains
changeAntibodies are glycoproteins belonging to the immunoglobulin superfamily; the terms antibody and immunoglobulin are often used interchangeably.[6] Antibodies are typically made of basic structural units—each with two large heavy chains and two small light chains. There are several different types of antibody heavy chains, and several different kinds of antibodies, which are grouped into different isotypes based on which heavy chain they possess. Five different antibody isotypes are known in mammals. They help direct the appropriate immune response for each different type of foreign object they encounter.[11]
Variable tips
changeThough the general structure of all antibodies is very similar, a small region at the tip of the protein is extremely variable, allowing millions of antibodies with slightly different tip structures, or antigen binding sites, to exist. This region is known as the hypervariable region. Each of these variants can bind to a different antigen.[1] This enormous diversity of antibodies allows the immune system to recognize an equally wide variety of antigens.[6] The large and diverse population of antibodies is generated by random combinations of a set of gene segments that encode different antigen binding sites (or paratopes), followed by random mutations in this area of the antibody gene, which create further diversity.[11][12] Antibody genes also re-organize in a process called class switching, which allows a single antibody to be used by several different parts of the immune system.
Examples of Antibodies
changeAntistreptolysin O: The Streptolysin Antibody
changeThe specific antibody that is produced to fight Group A streptococcus is called Antistreptolysin-O, also known as ASO.[13]
Detection of ASO via antistreptolysin-O titer or anti-deoxyribonuclease B (anti-DNase B) is a common way of testing for a group A streptococcal infection. These tests indicate if streptolysin antibodies are present. These proteins are only synthesized by lymphocytes in the event that streptolysin enters the immune system, thus this is a useful way of determining if a patient has the infection.
Medical Applications of Antibodies
changeUnderstanding the function and mechanism of antibodies is a powerful tool for disease treatment. Though most antibodies used in current clinical research are developed from mouse monoclonal antibodies, this method is allowing the scientific community to make great progress. Such developments include creating antibodies with highly conserved targets and the synthesis of new structures based on the antibodies themselves. There have also been advancements in increasing the binding affinity of antibodies to their associated antigens, a crucial quality to the effectiveness of the antibodies themselves.[14]
Antibodies are constantly being developed in order to protect the immune system from harmful pathogens. For example, a team of researchers is working on creating a monoclonal antibody that combats Human T-cell leukemia virus type-1 (HTLV-1). The antibody specifically targets the HTLV-1 envelope protein gp46 glycoprotein as a means of destroying HTLV-1-infected cells. The results of the study demonstrate that antibodies can be developed as a therapeutic tool against pathogen-infected cells, thereby serving as a means of treating diseases caused by said pathogens.[15]
Other websites
change- Animated depictions of how antibodies are used in ELISAArchived 2006-10-25 at the Wayback Machine and ELISPOTArchived 2006-10-12 at the Wayback Machine assays
References
change- ↑ 1.0 1.1 1.2 Janeway C.A. Jr.; et al. (2001). Immunobiology (5th ed.). Garland Publishing. ISBN 0-8153-3642-X. (electronic full text via NCBI Bookshelf).
- ↑ Litman G.W.; et al. (1993). "Phylogenetic diversification of immunoglobulin genes and the antibody repertoire". Mol. Biol. Evol. 10 (1): 60–72. doi:10.1093/oxfordjournals.molbev.a040000. PMID 8450761.
- ↑ For example, by blocking a part of a microbe that is essential for its invasion and survival.
- ↑ The system which produces antibodies in the blood plasma. Another system, cellular immunity, is done in the tissues by cells.
- ↑ Pier GB; Lyczak JB; Wetzler LM (2004). Immunology, infection, and immunity. ASM Press. ISBN 1-55581-246-5.
- ↑ 6.0 6.1 6.2 Rhoades RA, Pflanzer RG (2002). Human Physiology (4th ed.). Thomson Learning. ISBN 978-0-534-42174-8.
- ↑ "The immune system of the human body in defence against disease - Communicable diseases - AQA - GCSE Biology (Single Science) Revision - AQA". BBC Bitesize. Retrieved 2023-05-11.
- ↑ Mian I; Bradwell A; Olson A (1991). "Structure, function and properties of antibody binding sites". J Mol Biol. 217 (1): 133–51. doi:10.1016/0022-2836(91)90617-F. PMID 1988675.
- ↑ Fanning LJ; Connor AM; Wu GE (1996). "Development of the immunoglobulin repertoire". Clin. Immunol. Immunopathol. 79 (1): 1–14. doi:10.1006/clin.1996.0044. PMID 8612345.
- ↑ Nemazee D (2006). "Receptor editing in lymphocyte development and central tolerance". Nat Rev Immunol. 6 (10): 728–40. doi:10.1038/nri1939. PMID 16998507. S2CID 2234228.
- ↑ 11.0 11.1 11.2 Market E & Papavasiliou FN (2003). "V(D)J recombination and the evolution of the adaptive immune system". PLOS Biol. 1 (1): E16. doi:10.1371/journal.pbio.0000016. PMC 212695. PMID 14551913.
- ↑ 12.0 12.1 Diaz M & Casali P (2002). "Somatic immunoglobulin hypermutation". Curr Opin Immunol. 14 (2): 235–240. doi:10.1016/S0952-7915(02)00327-8. PMC 4621002. PMID 11869898.
- ↑ "Antistreptolysin O and Anti-Deoxyribonuclease B Titers". publications.aap.org. Retrieved 2023-05-11.
- ↑ Loo, LiNa; Robinson, Matthew K.; Adams, Gregory P. (2008). "Antibody engineering principles and applications". Cancer Journal (Sudbury, Mass.). 14 (3): 149–153. doi:10.1097/PPO.0b013e318173a5d5. ISSN 1528-9117. PMID 18536554. S2CID 30015983.
- ↑ Hatayama, Yasuyoshi; Yamaoka, Yutaro; Morita, Takeshi; Jeremiah, Sundararaj Stanleyraj; Miyakawa, Kei; Nishi, Mayuko; Kimura, Yayoi; Mitsunaga, Makoto; Iwase, Tadayuki; Kimura, Hirokazu; Yamamoto, Naoki (2022-09-29). "Development of a Monoclonal Antibody Targeting HTLV-1 Envelope gp46 Glycoprotein and Its Application to Near-Infrared Photoimmuno-Antimicrobial Strategy". Viruses. 14 (10): 2153. doi:10.3390/v14102153. ISSN 1999-4915. PMC 9608601. PMID 36298708.