Enzyme
An enzyme is a protein molecule in cells which works as a biological catalyst.[1] Enzymes speed up chemical reactions in the body, but do not get used up in the process, so they can be used over and over again.
Almost all biochemical reactions in living things need enzymes. With an enzyme, chemical reactions go much faster than they would without the enzyme.[2]p39 Other biocatalysts are catalytic RNA molecules, called ribozymes.
The substances at the start of a reaction are called substrates. The substances at the end of a reaction are the products. Enzymes work on the substrates, and turn them into products. The study of enzymes is called enzymology.
The first enzyme was found in 1833 by Anselme Payen.
Enzyme structure
changeThere are thousands of different enzymes and each one is specific to the reaction which it catalyses. Enzymes have names which show what they do. Enzyme names usually end in –ase to show that they are enzymes. Examples of this include ATP synthase. It makes a chemical called ATP. Another example is DNA polymerase. It reads an intact DNA strand and uses it as a template to make a new strand.
One example of an enzyme is amylase, found in saliva. It breaks down starch molecules into smaller glucose and maltose molecules. Another kind of enzyme is lipase. It breaks down fats into smaller molecules, fatty acids and glycerol.
The proteases are a whole class of enzymes. They break down other enzymes and proteins back into amino acids.[3] Nucleases are enzymes that cut DNA or RNA, often in specific place in the molecule.
Enzymes are not only for breaking large chemicals into smaller chemicals. Other enzymes take smaller chemicals and build them up into bigger chemicals, and do many other chemical tasks. The classification below lists the main types.
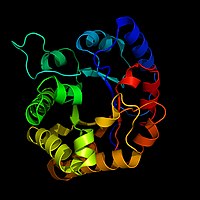
Biochemists often draw a picture of an enzyme to use as a visual aid or map of the enzyme. This is hard to do because there may be hundreds or thousands of atoms in an enzyme. Biochemists can not draw all this detail. Instead, they use ribbon models as pictures of enzymes. Ribbon models can show the shape of an enzyme without having to draw every atom.
Most enzymes will not work unless the temperature and pH are just right. In mammals the right temperature is usually about 37oC degrees (body temperature). The correct pH can vary greatly. Pepsin is an example of an enzyme that works best when pH is about 1.5.[4]
Heating an enzyme above a certain temperature will destroy the enzyme permanently. It will be broken down by protease and the chemicals will be used again.
Some chemicals can help an enzyme do its job even better. These are called activators. Sometimes, a chemical can slow down an enzyme or even make the enzyme not work at all. These are called inhibitors. Most drugs are chemicals that either speed up or slow down some enzyme in the human body.
Lock and key model
changeEnzymes are very specific. In 1894 Emil Fischer suggested that both the enzyme and the substrate have specific complementary geometric shapes that fit exactly into one another.[5] This is often referred to as "the lock and key" model. However, this model fails to explain what happens next.
In 1958, Daniel Koshland suggested a modification to the lock and key model. Since enzymes are rather flexible structures, the active site is reshaped by interactions with the substrate.[6] As a result, the substrate does not simply bind to a rigid active site. The amino acid side-chains of the active site are bent into positions so the enzyme does its catalytic work. In some cases, such as glycosidases, the substrate molecule also changes shape slightly as it enters the active site.[7]
Function
changeThe general equation for an enzyme reaction is:
- Substrate + Enzyme –> Substrate:Enzyme –> Product:Enzyme –> Product + Enzyme
Enzymes lower the activation energy of a reaction by forming an intermediary complex with the substrate. This complex is called an enzyme-substrate complex.
For example, sucrase, 400 times the size of its substrate sucrose, splits the sucrose into its constituent sugars, which are glucose and fructose. The sucrase bends the sucrose, and strains the bond between the glucose and fructose. Water molecules join in and make the cleavage in a fraction of a second. Enzymes have these key features:
- They are catalytic. They commonly increase the rate of reaction 10 billion-fold.[2]p39 The enzyme itself is not changed by the reaction.
- They are effective in tiny amounts. One enzyme molecule may convert 1000 molecules of substrate a minute, and some are known to convert 3 million in a minute.[2]p39
- They are highly specific. One enzyme will only carry out one of the many reactions of which a substrate is capable.
Control of enzyme activity
changeThere are five main ways that enzyme activity is controlled in the cell.
- Enzyme production (transcription and translation of enzyme genes) can be increased or reduced in response to changes in the cell's environment. This form of gene regulation is called enzyme induction and inhibition. For example, in bacteria which are resistant to antibiotics such as penicillin, enzymes are induced which hydrolyse the penicillin molecule.
- Enzymes can occur in different cellular compartments. For example, fatty acids are synthesized by one set of enzymes in the cytosol, endoplasmic reticulum and Golgi apparatus. Then they are used by a different set of enzymes as a source of energy in the mitochondria.[8]
- Enzymes can be regulated by their own products. For example, the end product(s) often inhibit one of the first enzymes of the pathway. Such a regulatory mechanism is called negative feedback, because the amount of the end product produced is regulated by its own concentration. This prevents the cells making too much enzyme. The control of enzyme action helps keep a stable internal environment in living organisms.
- Enzymes can be regulated by being modified after their production. An example is the cleavage of the polypeptide chain. Chymotrypsin, a digestive protease, is produced in inactive form in the pancreas and transported in this form to the stomach where it is activated. This stops the enzyme from digesting the pancreas or other tissues before it enters the gut. This type of inactive precursor to an enzyme is known as a zymogen.
- Some enzymes may become activated when they move to a different environment (e.g. from high pH to low pH). For example, haemagglutinin in the influenza virus is activated by a change in shape. This is caused by the acidic conditions which occur inside the host cell's lysosome.[9]
Enzyme inhibitors
changeInhibitors can be used to stop an enzyme from binding to a substrate. This may be done to slow down an enzyme-controlled reaction. The inhibitors fit loosely or partially into the enzyme's active site. This prevents or slows down an enzyme-substrate complex being formed.
Denaturation
changeDenaturation is the irreversible alteration of an enzyme's active site, caused by an extreme change in temperature or pH. It will decrease the rate of reaction because the substrate molecule will not be able to fit into the active site, so products cannot be formed.
Cofactors
changeCofactors, or coenzymes, are helper molecules which are needed to make an enzyme work. They are not proteins, and may be organic or inorganic molecules. Both types of molecules sometimes contain a metal ion at the centre, such as Mg2+, Cu2+, Mn2+ or iron-sulphur clusters. This is because such ions may act as electron donors, and this is important in many reactions. The need of enzymes for various little helpers is the basic reason why animals, including ourselves, need trace elements and vitamins.[10][11]
Classification
changeEnzymes have been classified by the International Union of Biochemistry. Their Commission on Enzymes has grouped all known enzymes into six classes:
- Oxido-reductases: catalyse transfer of electrons
- Transferases: move functional group from one molecule to another
- Hydrolases: add –OH (hydroxyl) group
- Lyases: split chemical bonds, and often add double bond or ring structure
- Isomerases: A –> B where B is an isomer of A
- Ligases: join two large molecules: Ab + C –> A–C + b
The individual enzymes are given a four-figure number which classifies them in the database.[12]p145[13]
Uses of enzymes
changeEnzymes are used commercially for:
- making baby food - pre-digesting food for babies
- softening the centres of chocolates
- biological washing powder - which contains protease enzymes to break down the grime and dirt. It breaks the large, insoluble molecules into small, soluble molecules. It works at a lower temperature, so less energy is needed (thermostable)
Related pages
changeReferences
change- ↑ Concise Oxford Dictionary.
- ↑ 2.0 2.1 2.2 Kornberg, Arthur (1989). For the love of enzymes: the odyssey of a biochemist. Harvard University Press. ISBN 0-674-30775-5.
- ↑ Pugh, Maureen Barlow (ed) 2000. Stedman's medical dictionary, 27th ed. Baltimore, Maryland, USA: Lippincott, Williams & Wilkins. p65
- ↑ "Effects of pH". Retrieved 2010-04-29.
- ↑ Fischer E. (1894). "Einfluss der Configuration auf die Wirkung der Enzyme" [Influence of configuration on the action of enzymes]. Berichte der Deutschen Chemischen Gesellschaft zu Berlin. 27 (3): 2985–93. doi:10.1002/cber.18940270364.
- ↑ Koshland D. E. (1958). "Application of a theory of enzyme specificity to protein synthesis". Proc. Natl. Acad. Sci. 44 (2): 98–104. Bibcode:1958PNAS...44...98K. doi:10.1073/pnas.44.2.98. PMC 335371. PMID 16590179.
- ↑ Vasella A, Davies GJ, Bohm M. (2002). "Glycosidase mechanisms". Current Opinion in Chemical Biology. 6 (5): 619–29. doi:10.1016/S1367-5931(02)00380-0. PMID 12413546.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Faergeman NJ, Knudsen J (April 1997). "Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling". Biochem. J. 323 (Pt 1): 1–12. doi:10.1042/bj3230001. PMC 1218279. PMID 9173866.
- ↑ Carr C.M. & Kim P.S. (April 2003). "A spring-loaded mechanism for the conformational change of influenza hemagglutinin". Cell. 73 (4): 823–32. doi:10.1016/0092-8674(93)90260-W. PMID 8500173. S2CID 43599939.
- ↑ Metzler, David E.; Metzler, Carol M. (2001). Biochemistry: the chemical reactions of living cells. Academic Press. ISBN 978-0-12-492540-3.
- ↑ Bugg, Tim (1997). Introduction to enzyme and coenzymec chemistry. Blackwell. ISBN 978-0-86542-793-8.
- ↑ King R.C. Stansfield W.D. & Mulligan P.K. 2006. A dictionary of genetics, 7th ed. Oxford.
- ↑ "Enzyme database". Archived from the original on 2006-04-04. Retrieved 2020-09-15.
{{cite web}}: CS1 maint: bot: original URL status unknown (link)